Ancient DNA, new field of research
2002/03/01 Andonegi Beristain, Garazi - Elhuyar Zientziaren Komunikazioa Iturria: Elhuyar aldizkaria
In this two-day meeting, the discoveries of each group were made known and the future of the research of the old DNA was analyzed. Among the participants was the professor and researcher of the Leioa campus of the UPV, Conchi de la Rúa, with whom we were able to have a conversation. He has explained to us the research that has been carried out in a unique world and its experience.
What is ancient DNA?

Most of the genetic material in an organism is stored in the cell nucleus as a chromosome. Chromosomes are made up of 3 billion nucleotides encoding between 50,000 and 100,000 genes. “However, most of the DNA chain is not a gene encoder and this neutral character makes DNA work for genetic research,” explains Conchi.
If we compare ancient DNA with the DNA of living beings, there are some notable differences. First, the amount of old DNA obtained in extractions is very low compared to that obtained from a live, between 0.1 and 1%. And secondly, the resulting DNA chain is fragmented into chains of 100 base pairs, while in living beings chains of thousands of base pairs can be obtained.
These differences are due to the degradation processes that occur in the samples. “The effects of oxidation and hydrolysis that occur after the death of the organism cause the rupture of the DNA chain, being of great importance the conservation of the organism for the quality of the samples in a dry environment and without oxygen,” he explains. For this reason, the age limit of ancient DNA stands at 100,000 years.
Mitochondrial DNA
Due to the small number of old DNA molecules recovered, most researchers have aimed to work with mitochondrial DNA in the case of the old DNA. Because nuclear DNA is unique in each cell, mitochondrial DNA is abundant. This abundance increases the survival and chances of DNA recovery.
In the case of mitochondria, the length of the DNA chain is 16,500 pairs of nitrogen bases, representing 0.0005% of the human genome. The DNA of human mitochondria and several vertebrates is sequenced and in this sequence there is an area called the control area. This equivalent zone in base 1.100 is neutral, so that mutations only appear under the influence of time and not by natural selection. In addition, the control point supports 90% of mitochondrial DNA mutations.

To these particularities we must add another: Mitochondrial DNA is only received from its mother. “Mitochondrial DNA is found in the tail of the sperm, and during fertilization of the egg the tail is left out,” says Conchi, “thus leaving out the male mitochondrial DNA.”
This means halving the area of study and facilitating the tracking of the genetic drift mark.
Initiation
The first ancient DNA research was conducted in 1984 with an animal that disappeared 150 years ago. An old DNA sequence was first obtained from the dry muscle tissue of an equus quagga. From this sequence, they demonstrated the evolutionary relationship between the quagga and today's closest relatives, the zebra.
A year later, in 1985, other researchers recovered DNA from the surface cells of mummies. Of the 110 selected mummies, 23 were better preserved and only two of them obtained DNA. This step was the beginning of the study of ancient human DNA.
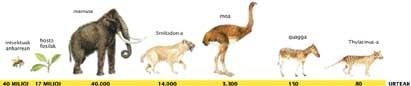
Continuing with the research, in the mid-1990s, fragments of DNA from missing vertebrates were sequenced, such as the Thylacinus or the Martsupial wolf (120 years), the Smilodón or tiger ‘with sabre teeth’ (14,000 years), the dark mammoth (10,000-50,000 years) and the model (3,300 years).
In addition, a magnolia leaf called Ma (17 million years) and several insects stored in amber were studied, but their studies are not currently considered reliable. “It has not been possible to repeat the results obtained in these laboratories or in the same or in others, which invalidates the results,” says Conchi. “However, it was demonstrated that ancient DNA was recoverable.”
In 1986, the new PCR analysis method was launched. This technique revolutionized DNA research and allowed the discovery of mitochondrial genes.
PCR method
The chain reaction or PCR method of polymerase consists of synthesizing millions of copies of a small fragment of DNA. In this cyclical process
Part of the DNA chain doubles exponentially until enough samples are available to apply the different analysis methods. PCR requires the design of short DNA chains with limiting function, complementary to the DNA chain being studied. The function of these limiters is to set the length of the DNA chain to duplicate, as they are added at the beginning and end. The enzyme called Taq polymerase then doubles the DNA chain.
Thus, the PCR method allowed expanding the studies to bones and teeth and, being a common material from archaeological excavations, opened the door to the study of the biological history of the centuries.
Methodological problems and pollution
However, the ability to duplicate the PCR method becomes a problem in the case of the old DNA. Ancient DNA has a high risk of contagion by coming into contact with any external element. Given this, and since the old DNA chains that are recovered are short, the PCR method has a high probability of copying external DNA fragments. Therefore, it is essential to adopt a series of measures to ensure the result of the dubbing.
Firstly, the physical isolation of the laboratory must be ensured so that it does not affect external elements; secondly, only the equipment intended for the recovery of old DNA must be used and finally the total sterility of the samples to be used must be ensured. These samples are usually extracted from the grains that are best preserved, usually using bones and teeth. “The amount of old available DNA extracted from the pulp is greater than that obtained from the rest of the tissues or bones, and also in the deposits is usually a common piece.”
In addition to all the measures mentioned, the studies must be repeated both in the same laboratory and in different laboratories, using more than one sample of the same specimen, which ensures the existence or not of contamination. Finally, to homologate the old DNA sequence, it must be compared with the universe of known sequences stored in the databases, subsequently analyzing the results in a suitable phylogenetic context and comparing them with the sequences of other living or dead species of the same taxonomic group.
What methods do you use for exams?
“Two methods can be used, one for sequencing and one for restrictive enzyme analysis.”

Sequencing is knowing the location of the nitrogen bases responsible for genetic information in the DNA chain. This multi-step automatic method consists of a special amplification of the DNA chain, where fluorescent fragments of different sizes are obtained. Then they are distributed by electrophoresis, that is, they separate each other in a gel on which an electric field is applied. Migration speed depends on size. In this process a laser reader detects the fluorescent emission of each part, which depends on the last nitrogen base that was added in the PCR process. Consequently, the nitrogen base is known according to the fluorescence that is released.
Restrictive enzymes analyze target sequences. “Meta sequences are short nitrogenous torque chains in the DNA chain.” Restrictive enzymes are able to break the DNA chain from where the target sequences are located. These meta sequences, which have a length of 4-6 pairs of nitrogen bases (pb), are different for each reducing enzyme, that is, a reducing enzyme must find a sequence of goals to break the DNA chain, which often does not exist. “Seeing whether or not the DNA chain reacts with different enzymes, we can know if it has different target sequences and whether or not it is target sequences, according to the different existing patterns, we can see relational genetics.” Methodologically, this second technique is simpler and faster, allowing for more positive results.
Next future
In recent years, ancient DNA research has focused on the search for phylogenetic relationships, the study of genetic variability over time, or the research of demographic changes. All this is due to advances in molecular biology and the development of the PCR technique. However, there are still obstacles such as the risk of contamination and poor conservation of old DNA.
Looking ahead, the study aims to improve the DNA extraction methodology and increase the number of old samples. In addition, new research will focus on deepening research into nuclear DNA. These indicators will allow you not only to analyze the information received from your mother, but also from your father. Will the future give us the limit of this science?
I Spain of ancient DNA. CongressIn the Residencia de la Vidriera of the Autonomous University of Madrid, located in Miraflores de la Sierra, the 1st Spanish Edition of ancient DNA was held. Congress. Among the guests were experts from 15 research centers dedicated to evolutionary biology, paleobiology, anthropology, molecular genetics, forensics and ictology. These groups of researchers set out to know and deepen the common objectives of their science. These objectives included the creation of a working group on ancient DNA. This group would establish a basic procedure to ensure the quality of research in this science. Another objective was to strengthen relations between laboratories, promoting collaboration between research groups. Finally, the intention was commented to publicize the potential applications of this scientific field so that people take into account the possible applications of ancient DNA research in other fields. |
What's in Euskal Herria?
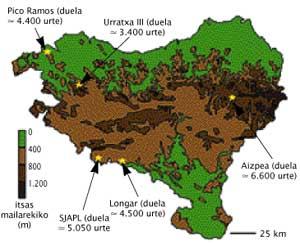
Conchi de la Rúa and his team direct their research to anthropology. The method of restrictive enzymes has allowed to classify 97% of pollution-free samples from various Basque deposits. Although the percentage seems high, it must be said that only 25% of the recovered samples are represented, since only the teeth in perfect condition have been analyzed to avoid the risk of contamination.
Among the findings, data have been obtained to oppose the theories of some Italian researchers. These Italian researchers reported on a group that between 10,000 and 15,000 years ago spread from the Basque Country to Europe. The aim of this theory was to explain the abundance of a genetic group currently existing in Basque society, the so-called V Haplotalde. The work of the Basque researchers has shown that there is the absence of this group in the old DNA samples, that is, there is no indication of that haplotalde era. What does this mean? “That this group of Haplots was not created here because if it were from here it should appear in samples less than 10,000 years old.”
Conchi and his companions put the view on genetic drift to explain the abundance of this group. “The mutation that defines the Haplotaldea could reach a high rate in a population group, as is the case of Gipuzkoa, at a time when the population was reduced, and subsequent population growth allowed to expand this mutation.”

Gai honi buruzko eduki gehiago
Elhuyarrek garatutako teknologia